How to browse and search#
Browse
- An intuitive “Google Maps” display enables easy navigation through the PD map.
- Clicking any element in the PD map (e.g., proteins, molecules or phenotypes) provides detailed annotations and links to external databases, which are displayed in the left panel (Fig. 3)presumably SERCH GENERIC is active. Otherwise, the information is visible on the bottom of the left panel (see Fig. 11: CREPPB protein).
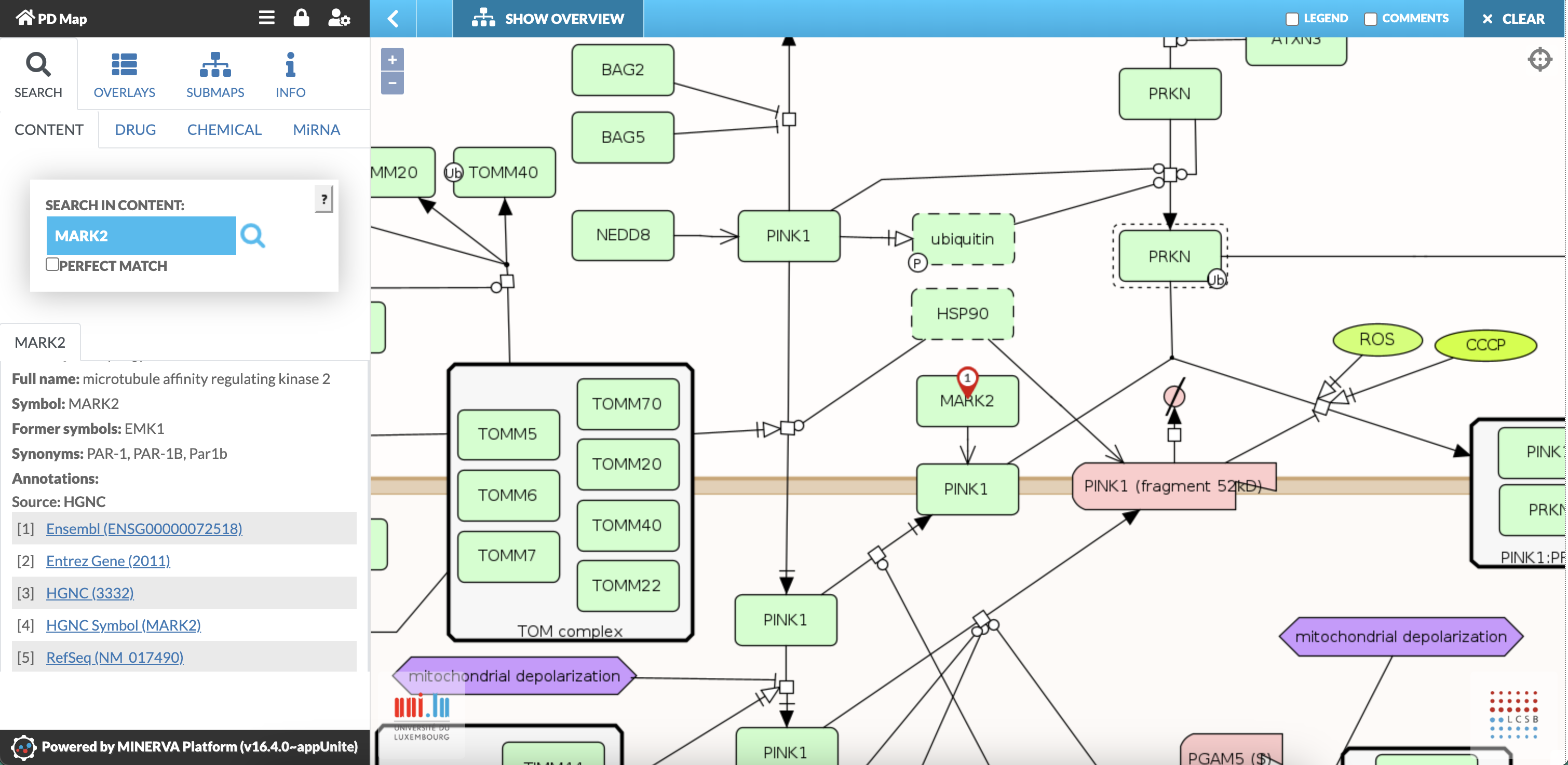
Fig. 3: Screenshot showing annotations of the protein: MARK2 (microtubule affinity-regulating kinase 2)
- By clicking on the bubble pinned to the element (Fig. 4A red arrow) additional information is displayed in a pop up box.
- For genes, miRNAs and proteins a special pop up box enables the search for interacting drugs, chemicals and miRNAs. First click on the element, then on the coloured number above and then the related checkbox (“Show all”) (Fig. 4).
- For more information on the different tools on drugs, chemicals and miRNAs interaction see related chapters 2.3, 2.4, and 2.5.
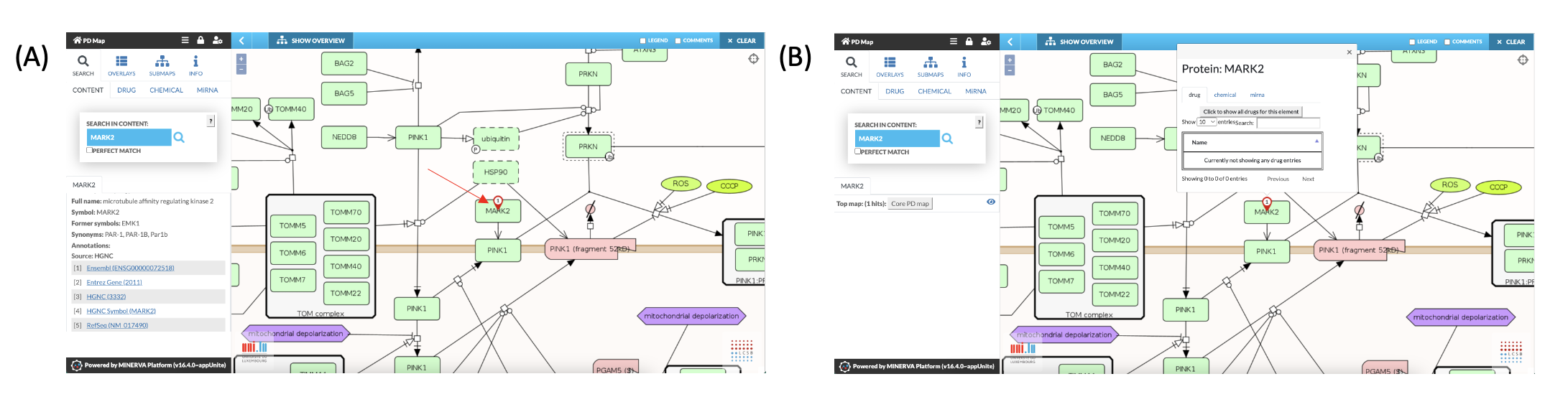
Fig. 4: Additional information and functions in relation to MARK2 displayed as pop up.
- Clicking on a connection line between elements (Fig. 5) provides detailed information on the underlying reaction (coloured in blue), displayed in the left panel. This includes information on reactants and links to the underlying sources (i.e., PubMed, Reactome or KEGG).
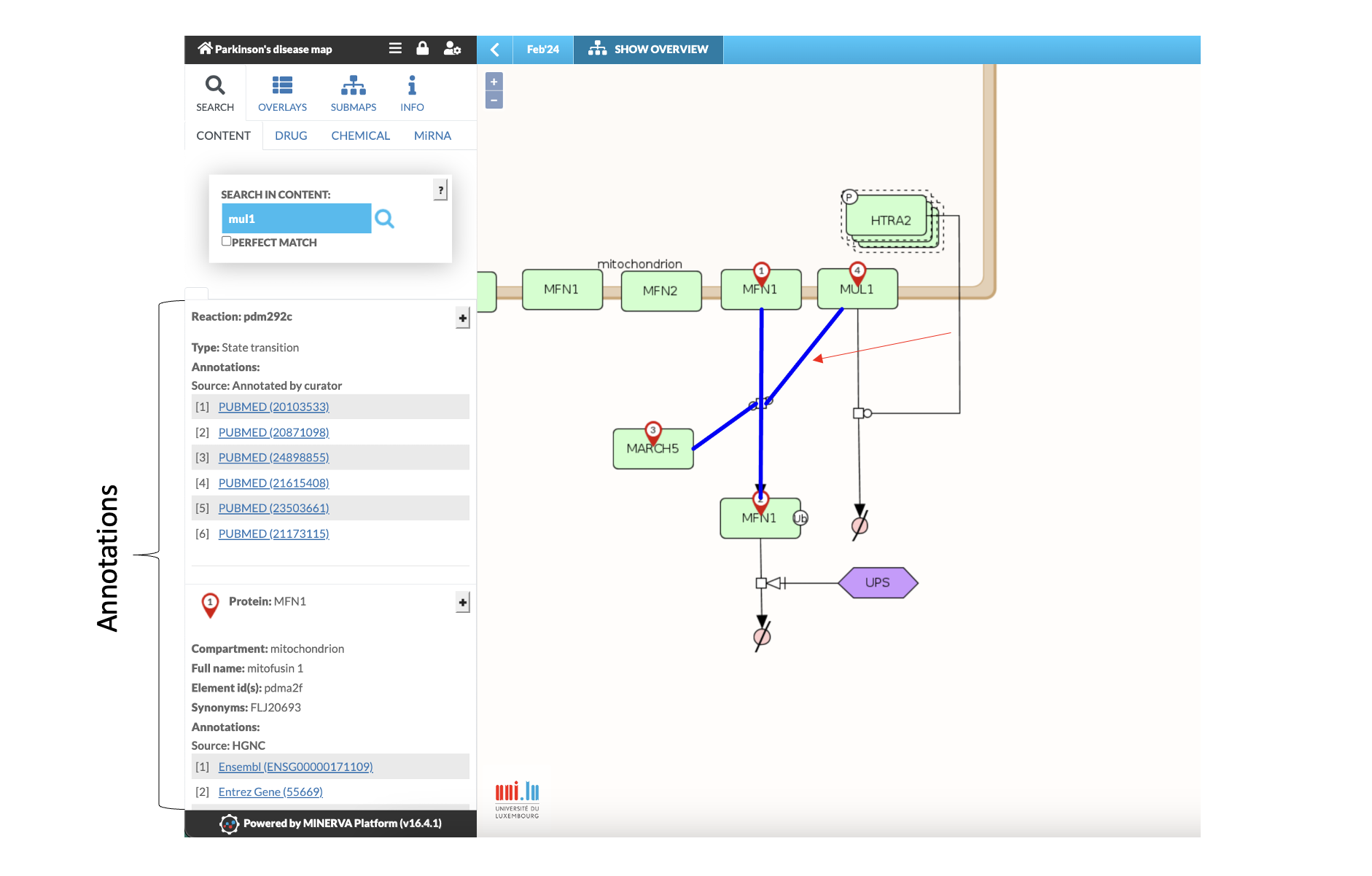
Fig. 5: Screenshot showing annotations of reaction pdm292c: MUL1 and MARCH5 mediate the ubiquitination of MFN1. Activated reaction is coloured in blue.
Search: CONTENT
- The “CONTENT SEARCH” function is located in the upper part of the left panel
- All elements (e.g., genes, proteins, small molecules, complexes) are searchable
- Search function also works with synonyms
- Search for multiple elements: separate names by semicolon (see Fig.6)
- Clicking on “PERFECT MATCH” reduces the search to those elements that exactly correspond to the input
- The location of the searched species is displayed on the PD map by coloured circles.
- Results from searches for multiple elements are displayed with different colours.
- The left panel provides annotations on the elements and the reaction types including links to external databases.
- Search for reactions: type in:reaction: reXXXX (XXXX = reaction number)
- Search for publications and related interaction: type pubmed:XXXXXXX (where XXXXXXX = PubMed ID).
- Search for publications also works with the specific publication list. Under “INFO” you find the link to the list of the current publications curated into the PD map. This list is fully searchable and contains links back to the PD map.
- Clicking on “CLEAR” (right upper corner) will remove all search results from the PD map
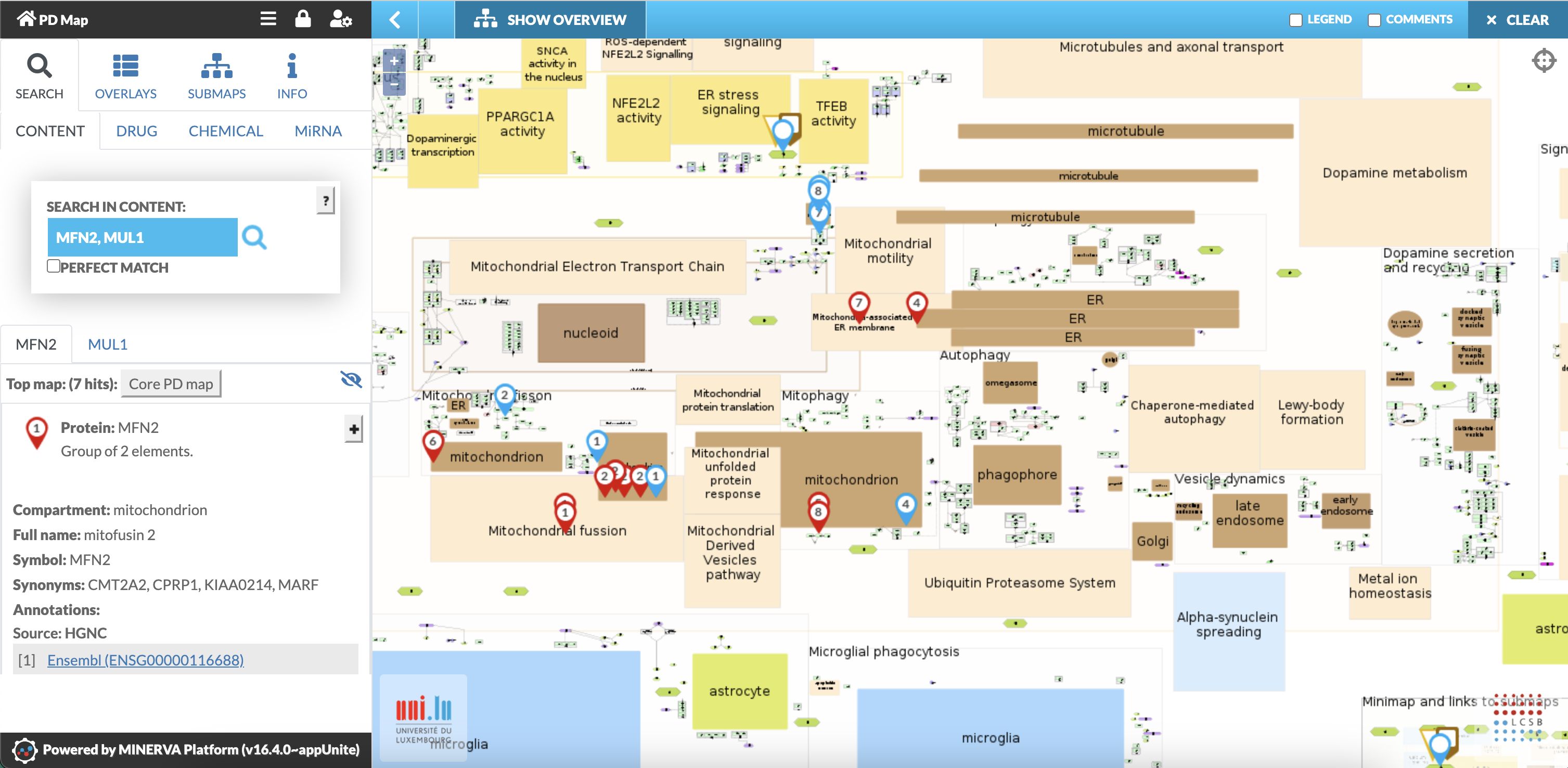
Fig. 6: Search for Mitofusin2 (MFN2) and Mulan1 (MUL1): All locations of MFN2 (red circle) and MUL1 (blue circle) are displayed in the PD map. Left panel shows annotations.
Search: How to use drug interface
- The “DRUG” interface allows to search for potential drug targets within the PD map.
- Activate the “DRUG” tab under “SEARCH”.
- Type drug names (synonyms or brand names also work) in the search field.
- Separate multiple search by semicolon
- Potential targets are fetched from the drug databases DrugBank and ChEMBL, displayed and marked in the PD map by numbers within a coloured rectangle.
- Additional information concerning the drugs and the potential targets, including links to drug databases and PubMed is provided in the left panel.
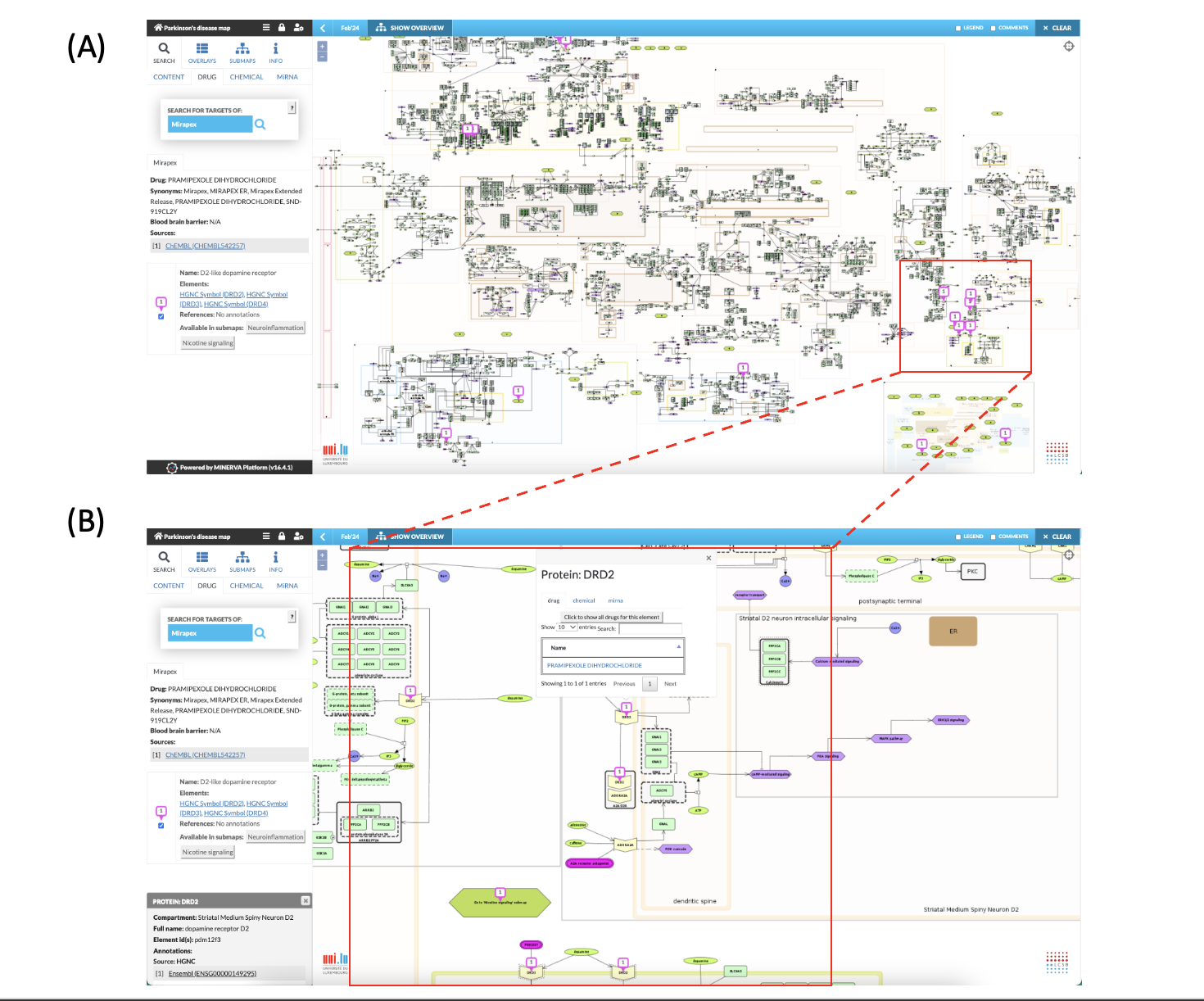
Fig. 7: Search for the drug “Mirapex”. (A) Information panel displays information on Pramipexole as Mirapex is a brand name for the drug Pramipexole. Drug targets are displayed in the map. (B) Pramipexole targets dopamine receptor (DRD2). The bubble pinned to the target (DRD2) provides information on drug interaction and offers additional functionalities to search for other drugs, chemical or miRNA that interacts with the DRD2 protein (see also Fig. 4B, Fig. 8, Fig. 9).
Please note:
- If within a multiple search several drugs hit the same element, the bubbles will overlap and only the one from the second search is visible on that element (see Fig. 8)
- The same happens when search queries from different tools targeting the same element in the PD map. Only the bubble from the first search is visible on the element that is targeted by the two or more search queries.
- All results are always displayed in the left panel.
- All results are displayed simultaneously in a pop up information field when clicking on the bubble pinned to the target (see Fig. 4; Fig. 7C, Fig. 8 and Fig 11).
- At the end of the list, the left panel shows also targets that are not in the PD map (see Fig.9).
Search: How to use chemical interaction interface
- The “CHEMICAL” interface allows to search for potential interactions of chemicals with the PD map elements based on the Comparative Toxicogenomics Database.
- Only interactions that are annotated to the disease term Parkinson’s disease (MeSH ID: D010300) in the CTD database are considered.
- Activate “CHEMICAL” tab under “SEARCH”.
- Type in the full name of a chemical according to the ctdbase/chem database; abbreviations or synonyms do not work (e.g., MTPT does not work but 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine).
- If the chemical name includes comma(s), place a semicolon behind the name to avoid a segmentation of the name during processing.
- Always separate multiple search by semicolon.
- Potential targets are fetched from the Comparative Toxicogenomics Database, displayed and marked in the PD map by a number within a coloured circle.
- Additional information concerning the chemical and the potential targets, including links to CTD databases and PubMed are provided in the left panel.
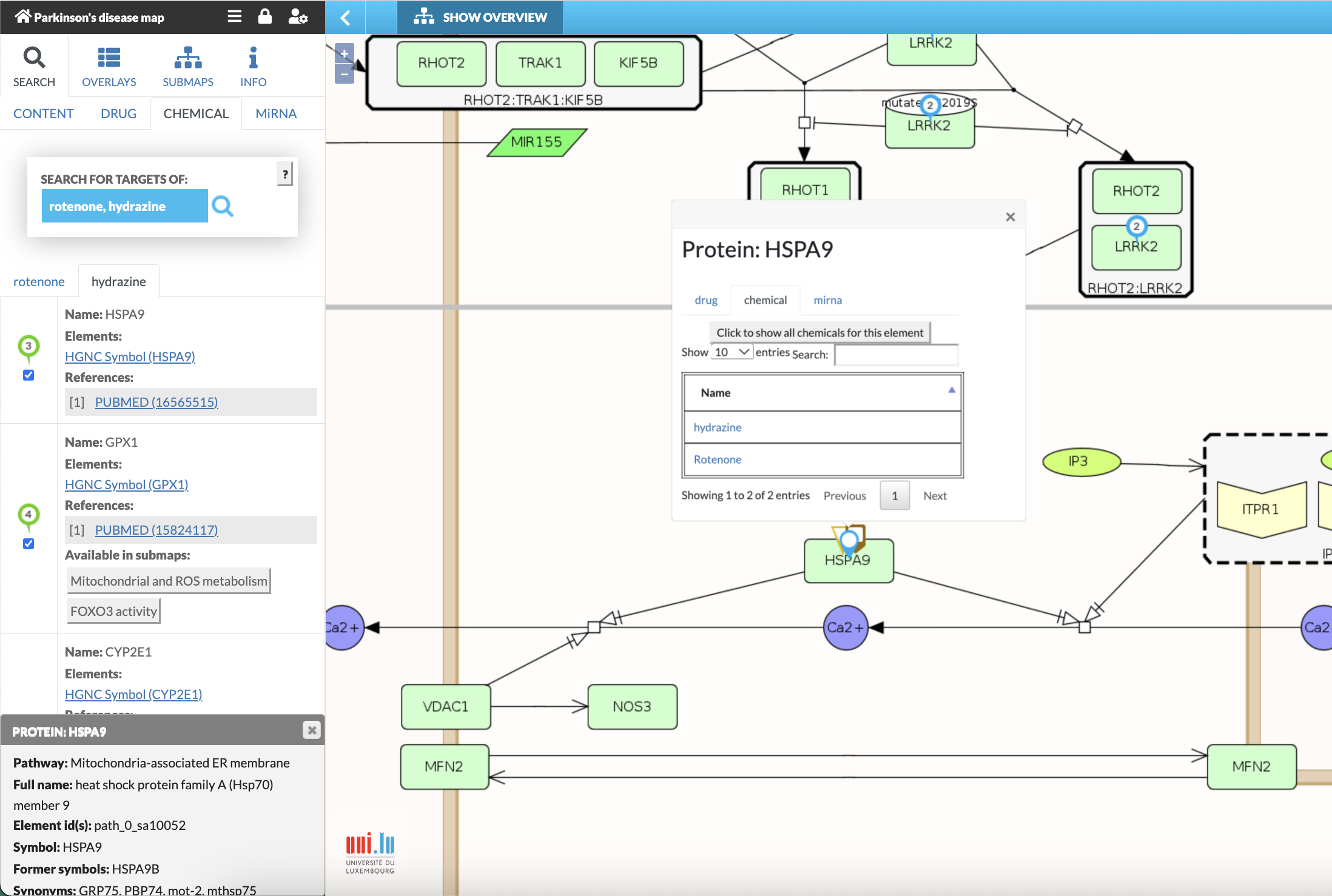
Fig. 8: Search for the interaction targets of “rotenone” and “hydrazine”. Information panel displays information on “hydrazine”. Interacting species are labelled in the map (green bubbles for hydrazine; blue bubbles for rotenone; multi-icon symbols for targets of both chemicals). Clicking on framed number above the target (e.g., green framed “3”) provides information on the target (i.e., HSPA9) and chemical interactions and offers additional functionalities to search for other drugs, chemical or miRNA that interacts with the target (see also Fig. 4; Fig. 7C, and Fig 11). Please note: If within a multiple search several drugs hit the same element, the bubbles will overlap and only the one from the second search is visible on that element.
Search: How to use miRNA interface
- The “MiRNA” interface allows to search for potential targets of miRNA within the PD map elements based on the miRNA database: miRTarBase.
- Only target interactions that have strong evidence according to miRTarBase criteria (i.e., reporter assays, western blot or qPCR analysis) are considered.
- Activate “MiRNA” tab under “SEARCH”.
- Type in the miRNA ID, use only mature sequence IDs e.g., hsa-miR-125a-3p (see:www.mirbase.org).
- Separate multiple search by semicolon.
- Matching targets from the miRTarBase are displayed in the PD map by pinned bubbles (here: coloured triangles).
- Additional information concerning the miRNAs and the potential targets, including links to miRTarBase and PubMed is provided in the left panel.
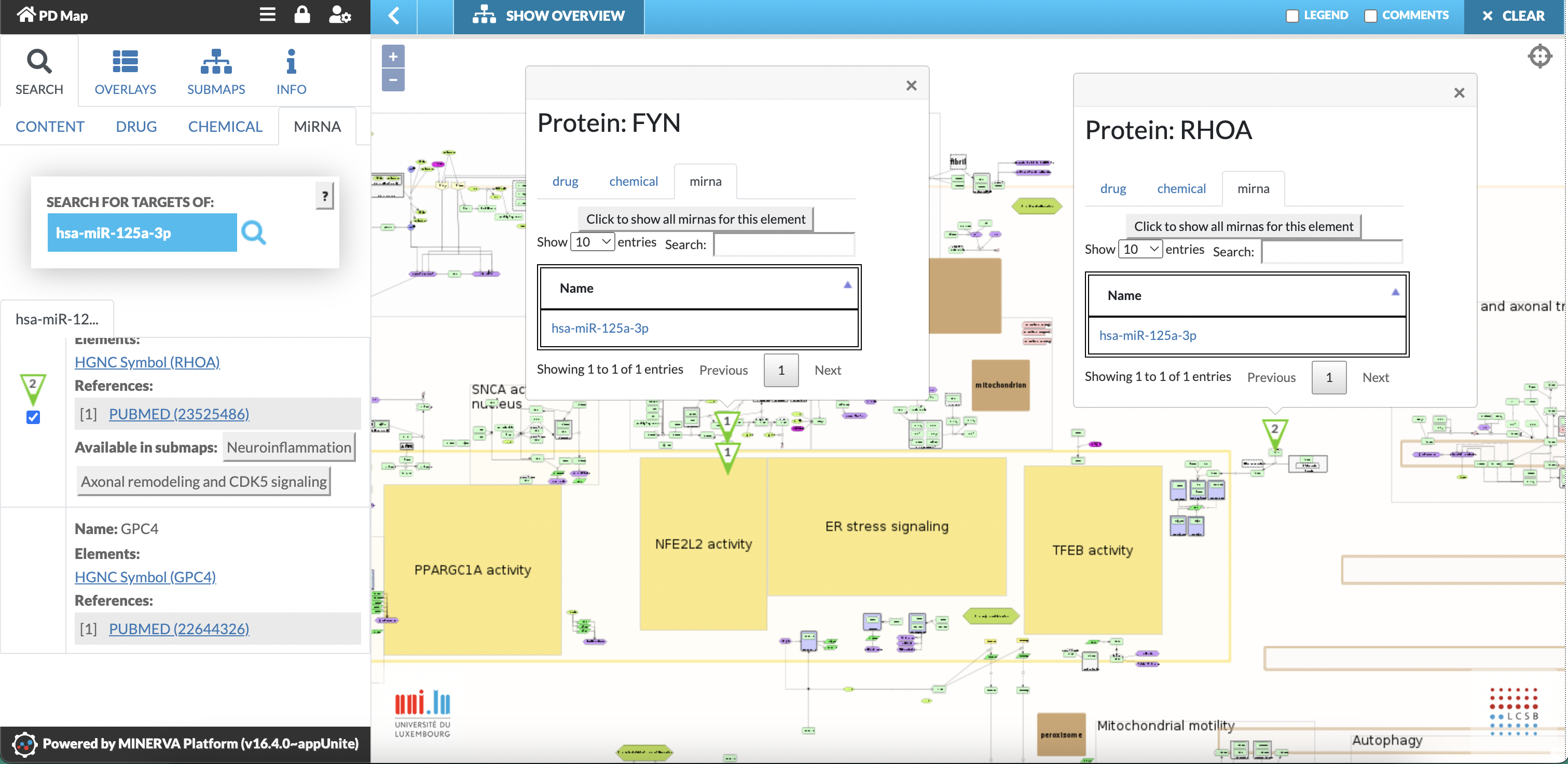
Fig. 9: Search for targets of hsa-miR-125a-3p. Left panel displays information on hsa-miR-125a-3p and its targets. Targets are marked by pinned bubbles (green triangles) in the PD map. Clicking on the pinned bubble provides information on miRNA interactions and offers additional functionalities to search for other miRNA, drugs, chemical that interacts with the target. Left panel shows also targets that are not in the PD map (here: GPC4).